Results
Movies
Molecular Dynamics
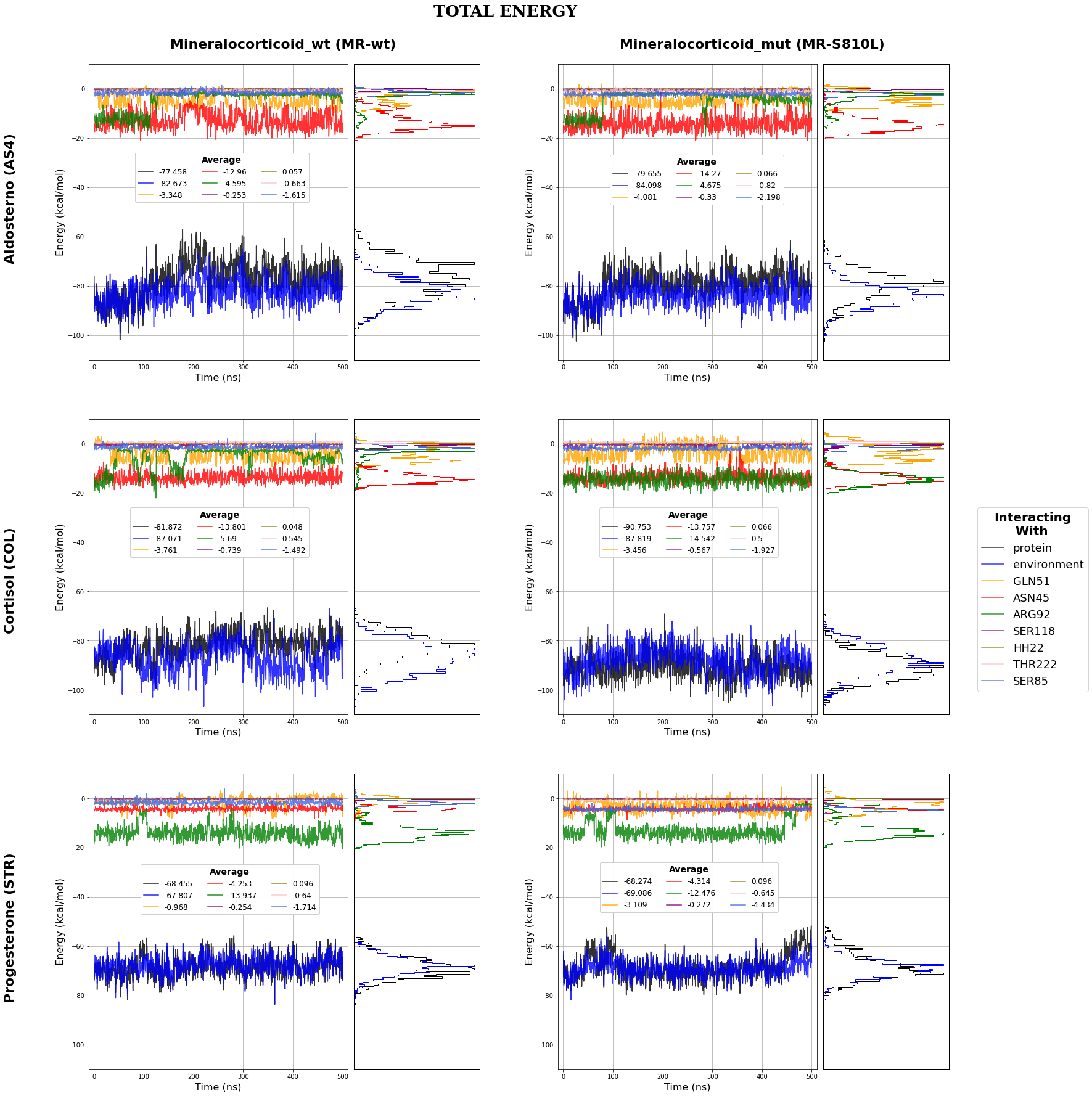
MR-AS4 MD simulation
MR-COL MD simulation
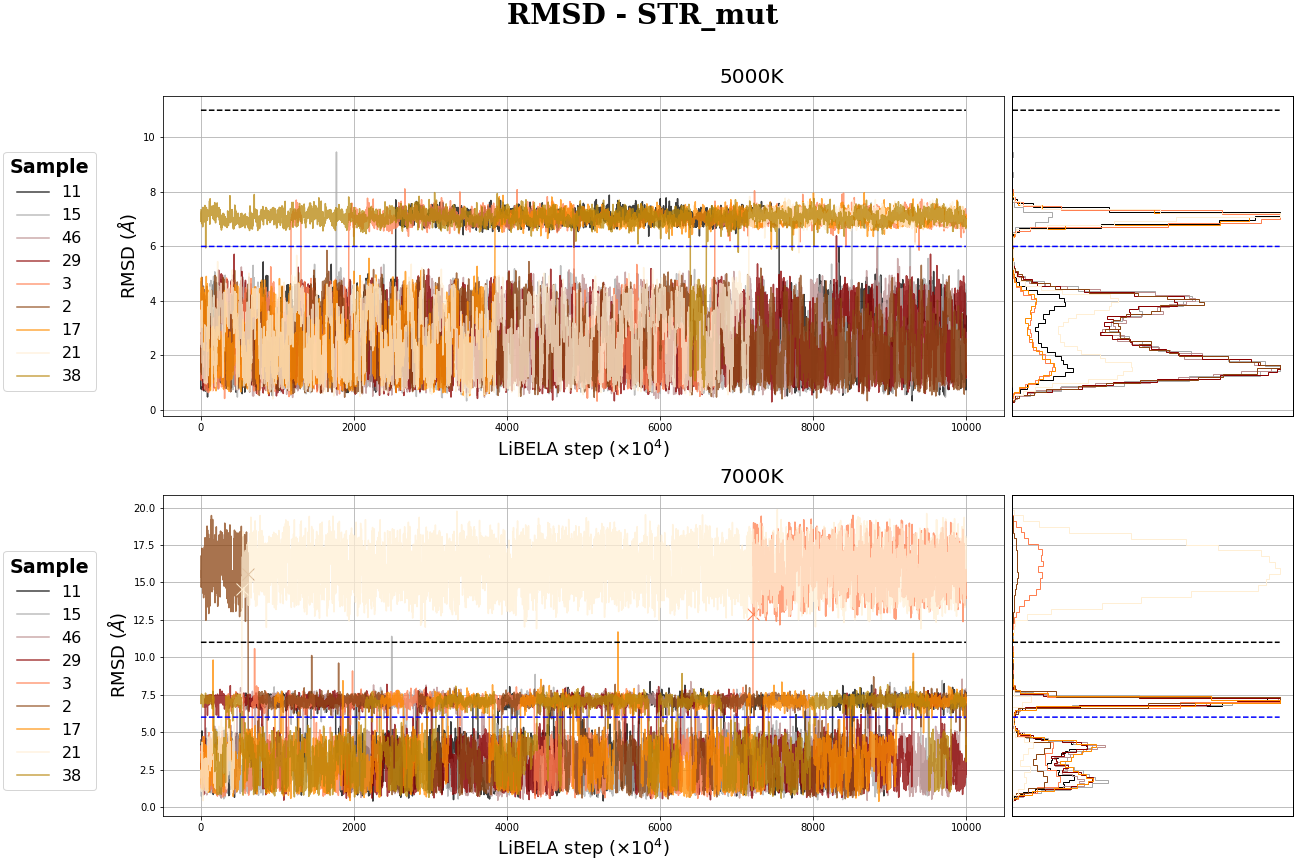
MR-STR MD simulation
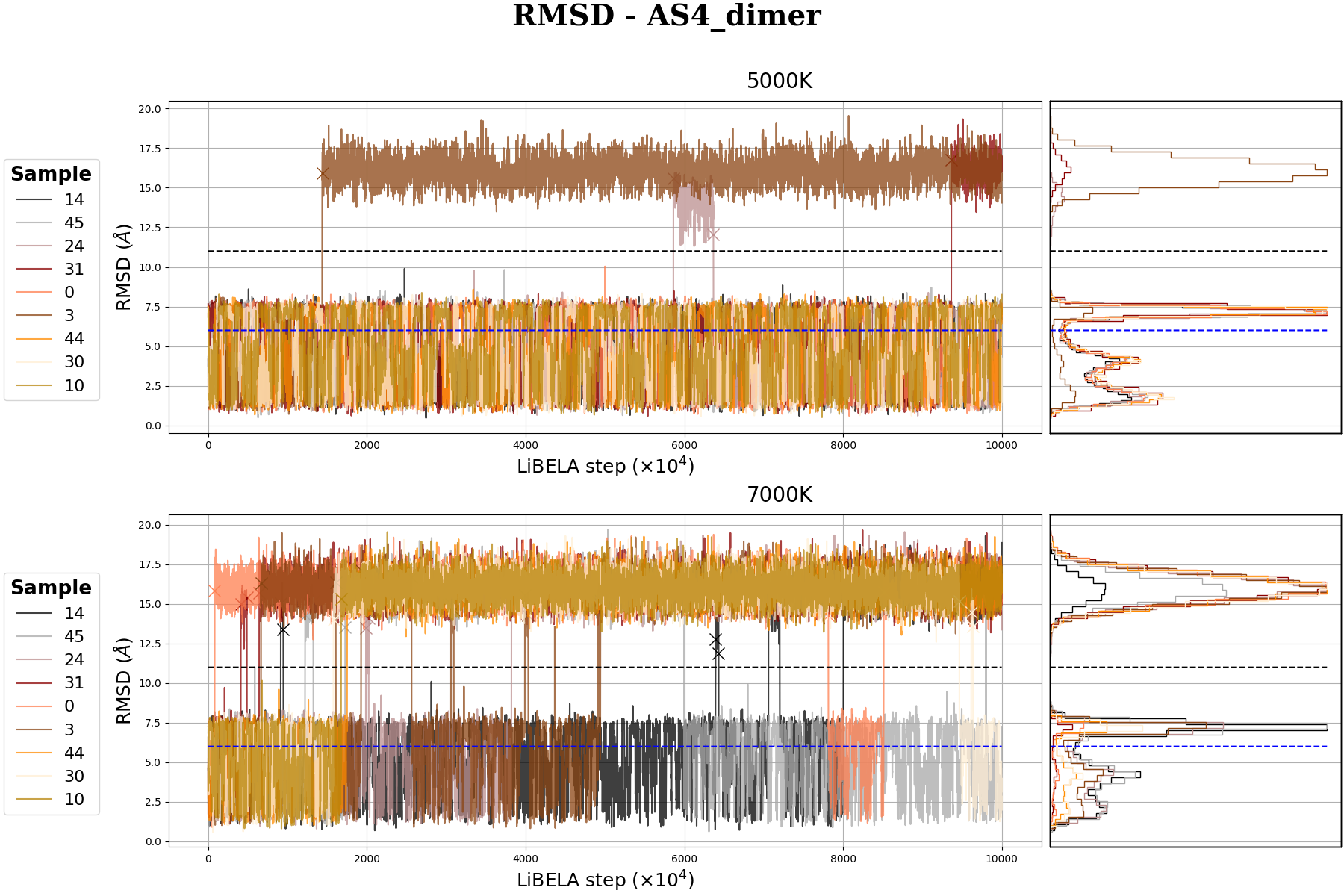
MRdimer-AS4 MD simulation
Monte Carlo
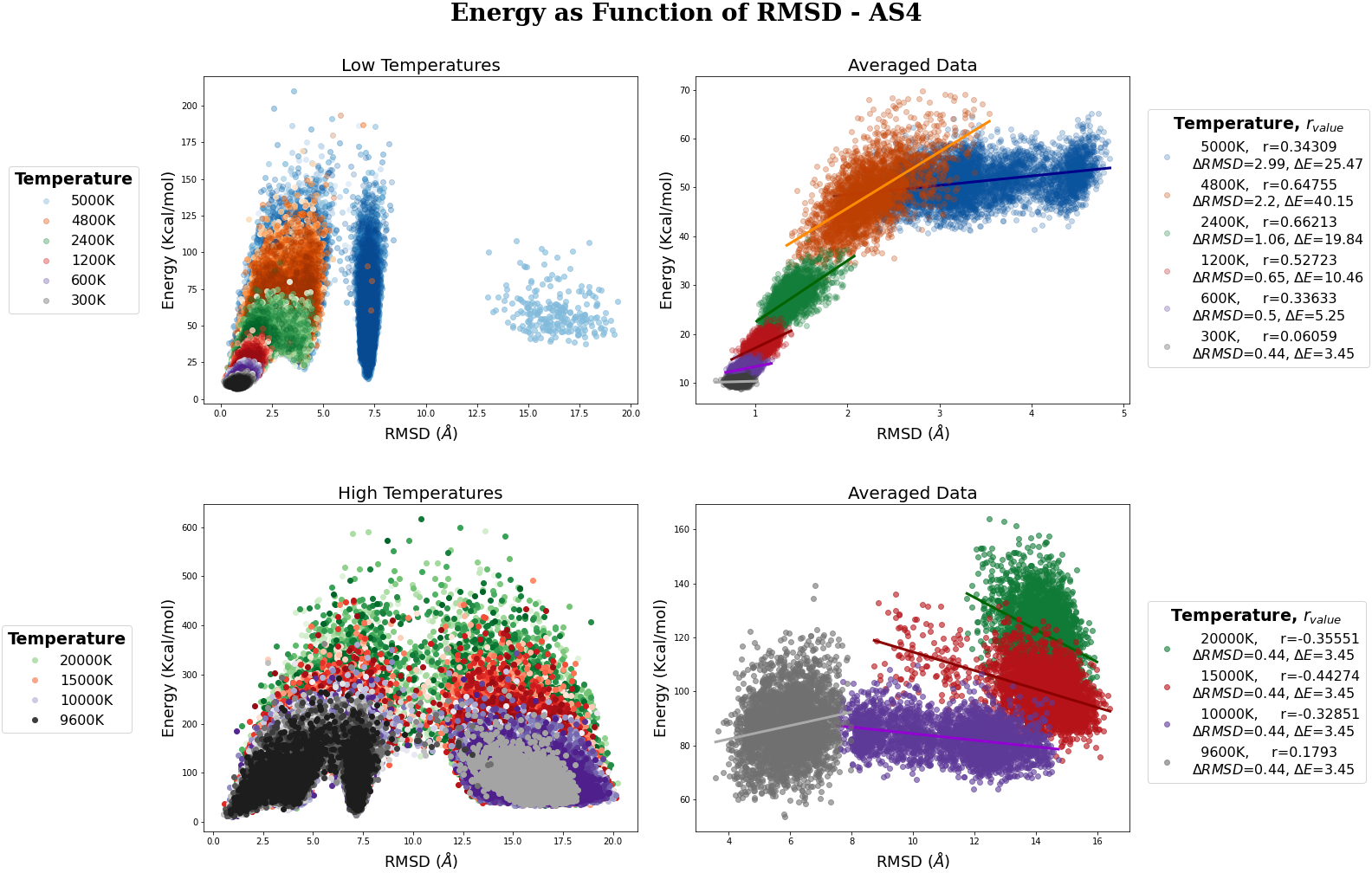
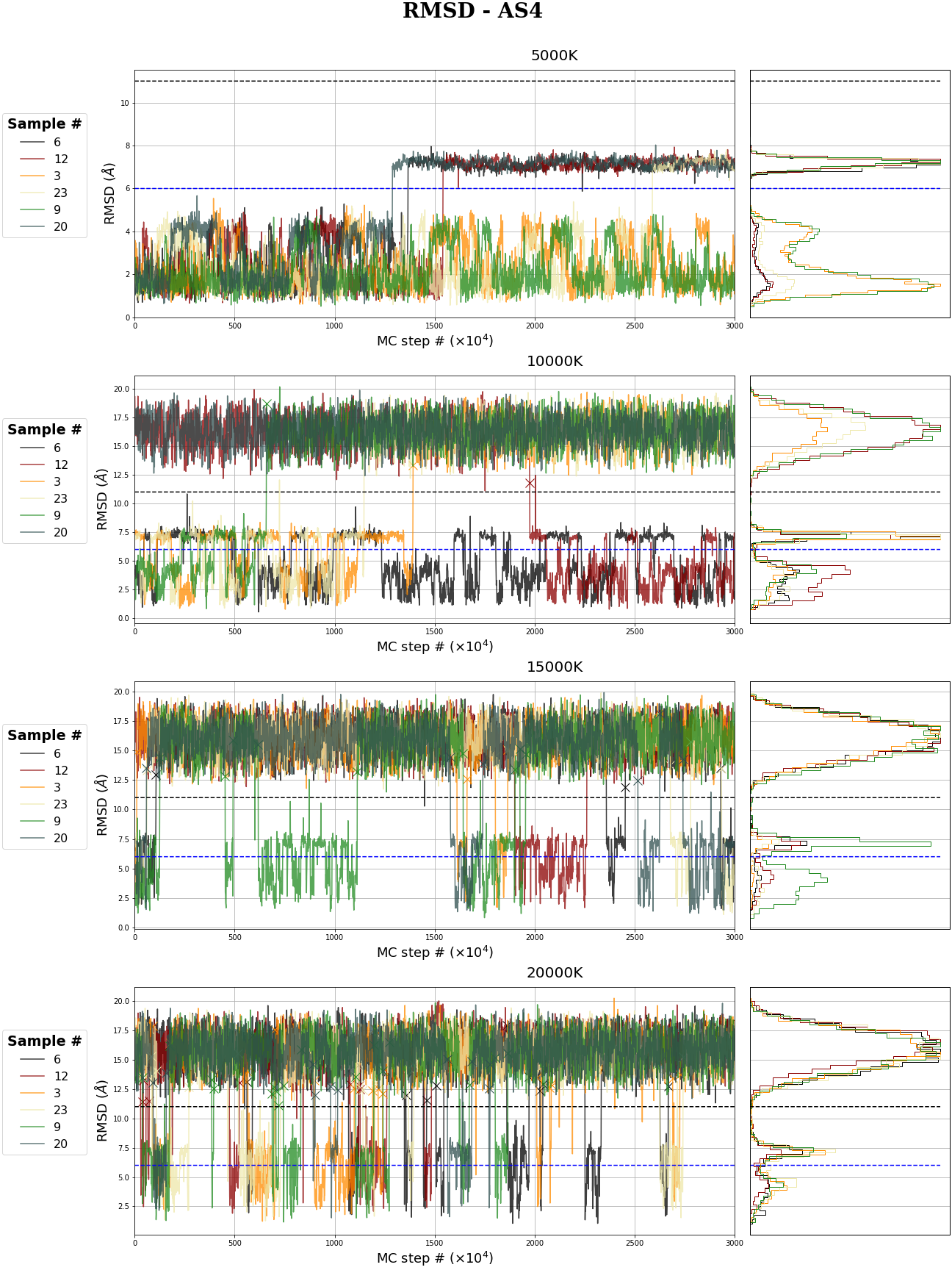
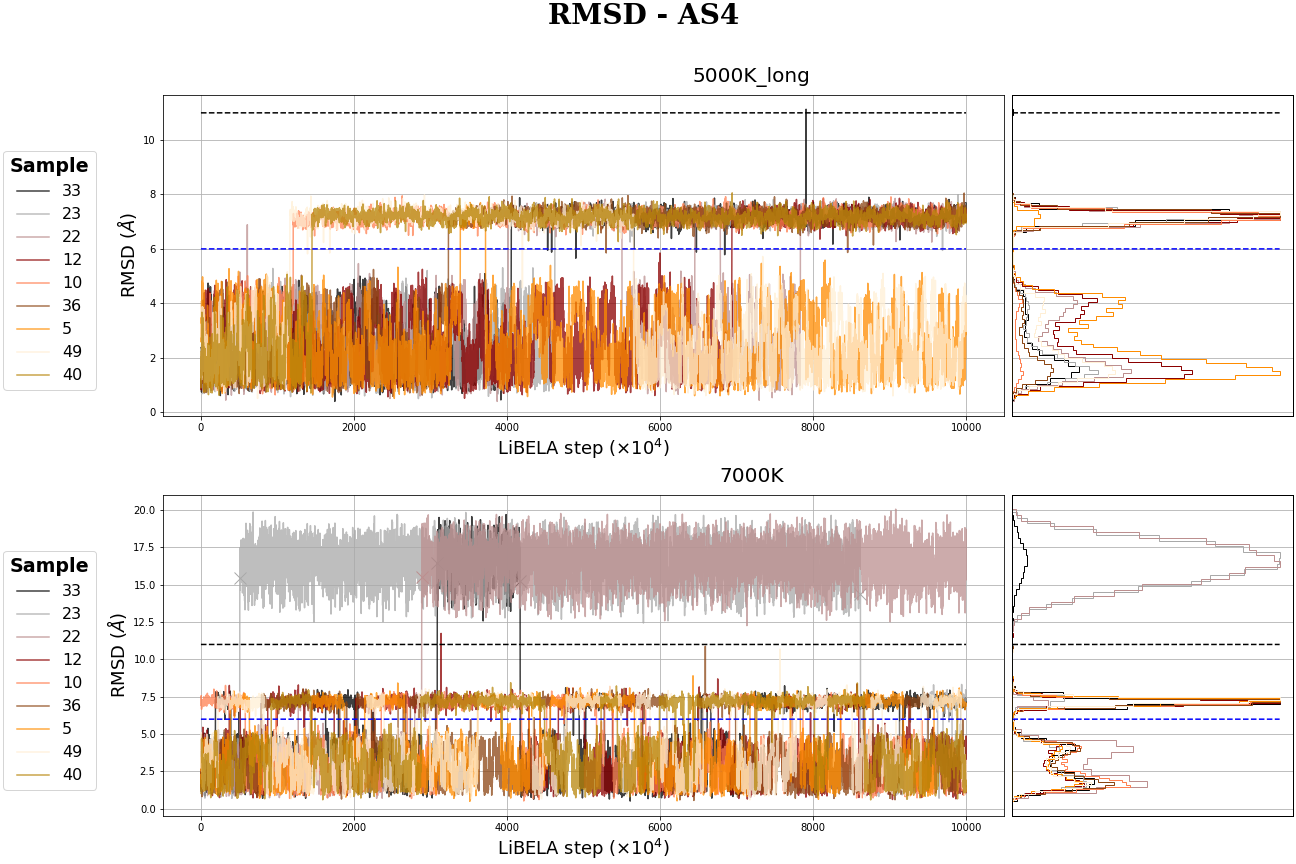
MR-AS4 MC simulation
Images
Molecular Dynamics
Monte Carlo
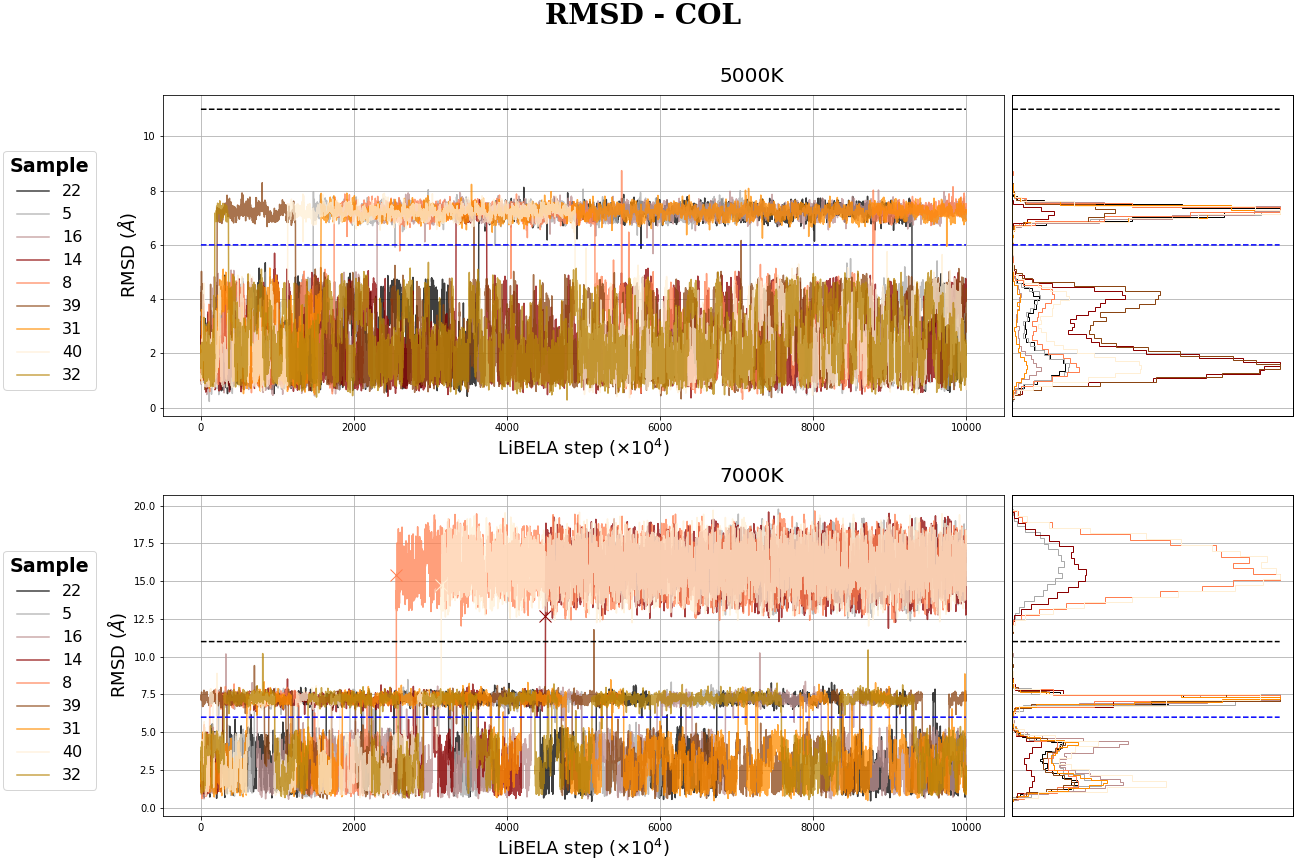
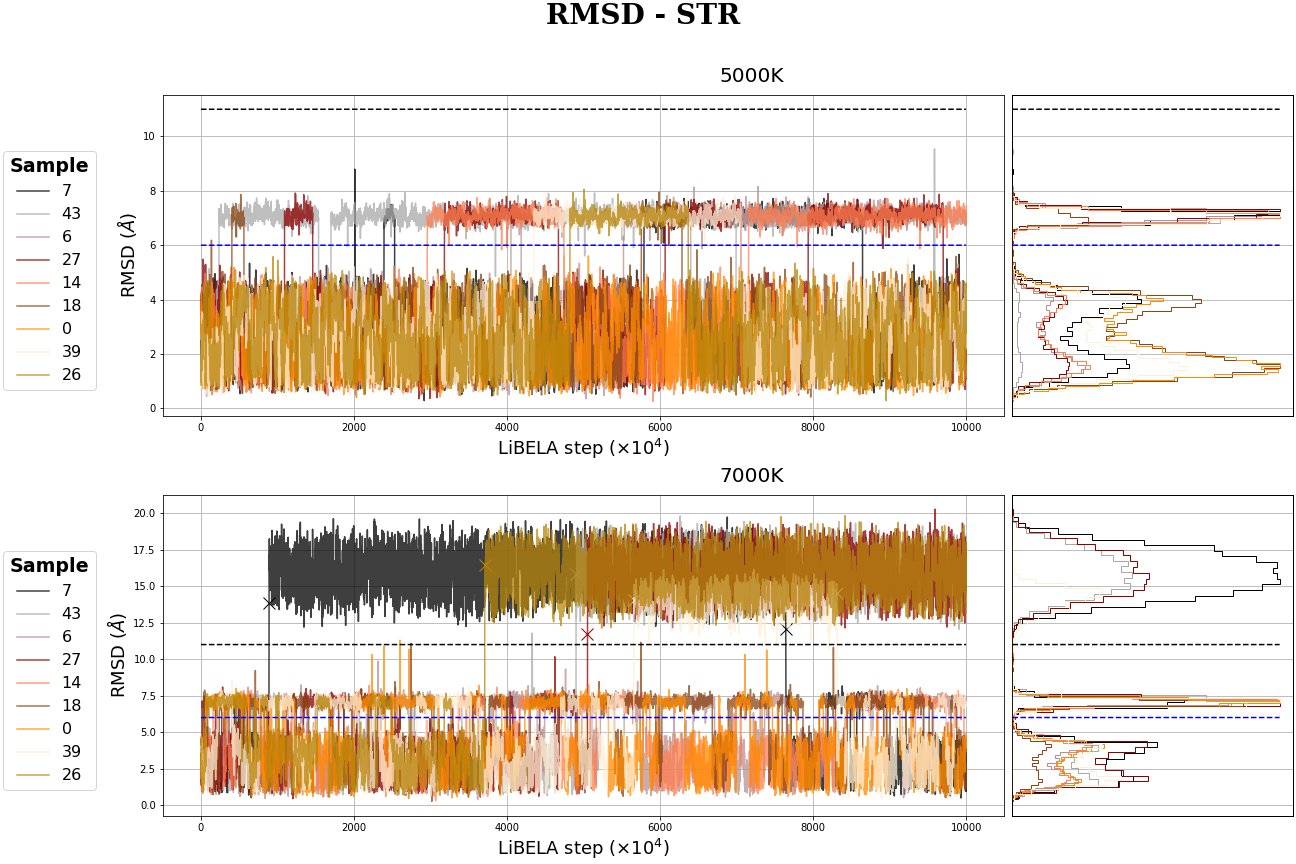
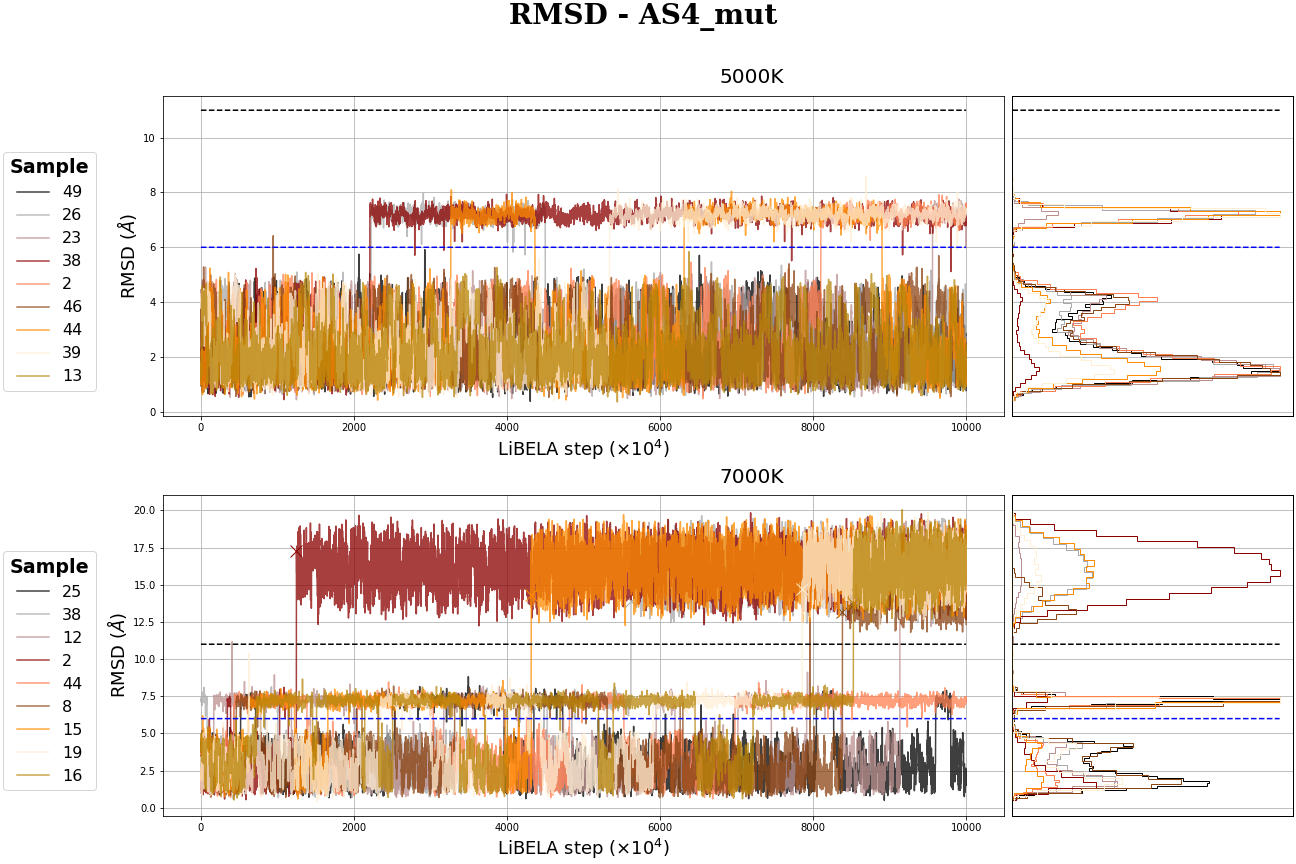
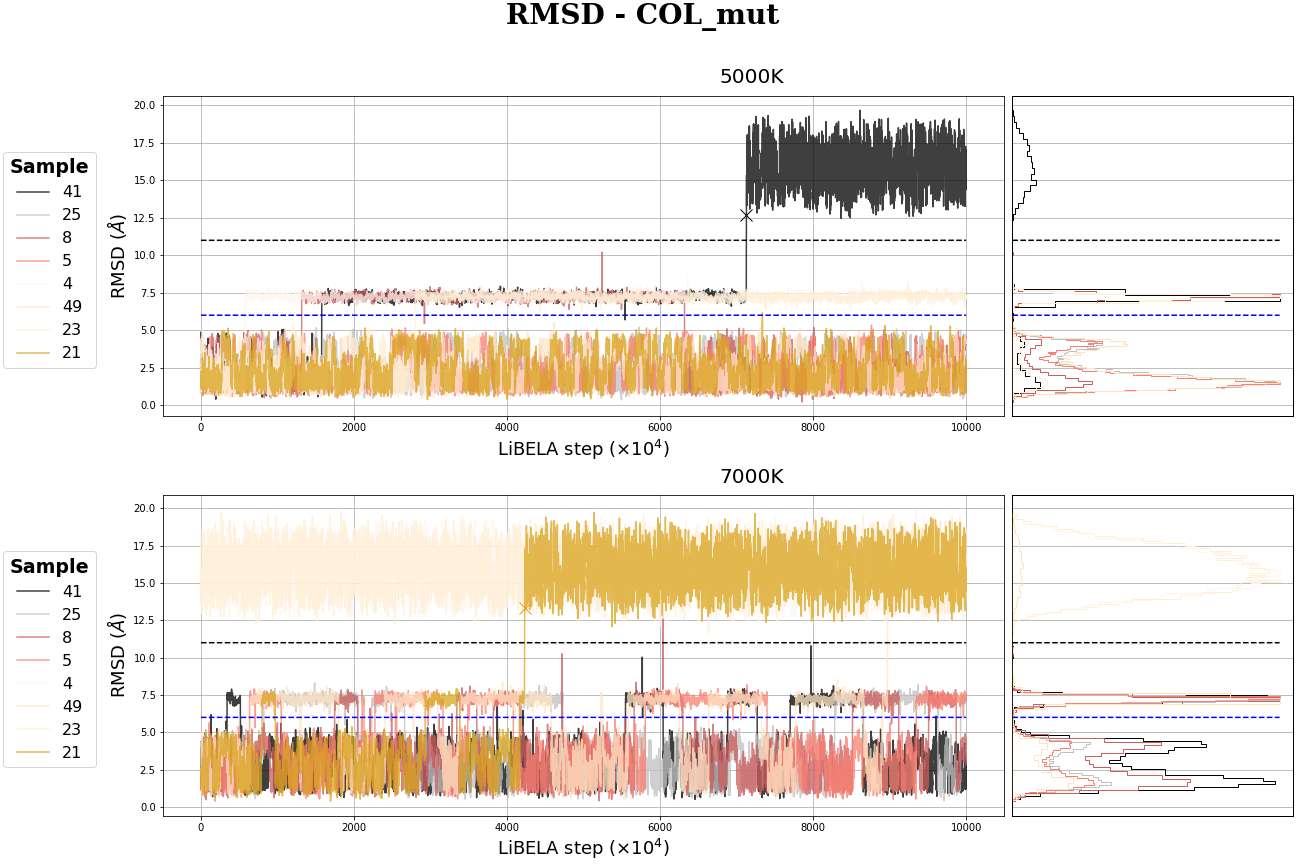
MC Table
| Ligand | Temperature (K) | Unbinding | Binding | Unbiding Probability (%) | Biding Probability (%) |
|---|---|---|---|---|---|
| AS4 | 5000K | 0 | 1 | 0.00 | 1.96 |
| AS4 | 7000K | 2 | 20 | 3.92 | 39.22 |
| COL | 5000K | 0 | 0 | 0.00 | 0.00 |
| COL | 7000K | 0 | 28 | 0.00 | 54.90 |
| STR | 5000K | 0 | 0 | 0.00 | 0.00 |
| STR | 7000K | 4 | 30 | 7.84 | 58.82 |
| AS4mut | 5000K | 0 | 3 | 0.00 | 5.88 |
| AS4mut | 7000K | 2 | 30 | 3.92 | 58.82 |
| COLmut | 5000K | 0 | 3 | 0.00 | 5.88 |
| COLmut | 7000K | 0 | 14 | 0.00 | 27.45 |
| STRmut | 5000K | 0 | 0 | 0.00 | 0.00 |
| STRmut | 7000K | 5 | 19 | 11.76 | 37.25 |
| AS4dimer | 5000K | 6 | 19 | 11.76 | 37.25 |
| AS4dimer | 7000K | 92 | 124 | 180.39 | 243.14 |
All simulation have 51 samples.
PyEMMA
A better aproach to analyse these data is using PyEMMA, a Python library for the estimation, validation and analysis Markov models of molecular kinetics and other kinetic and thermodynamic models from molecular dynamics (MD) data.
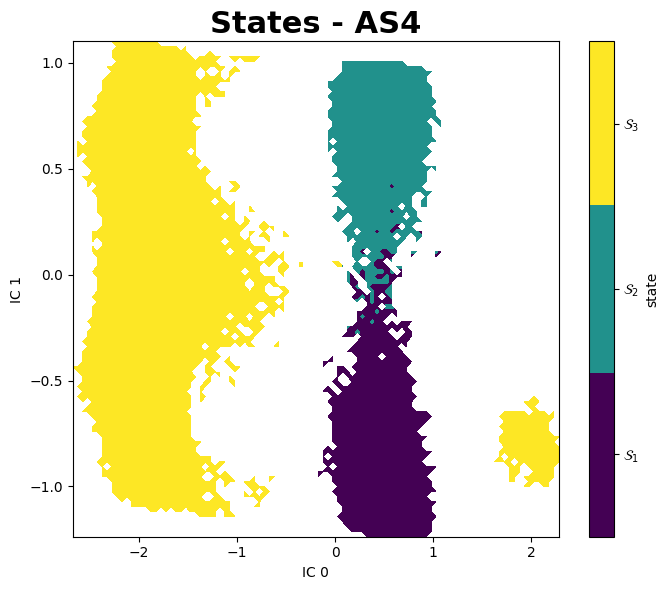
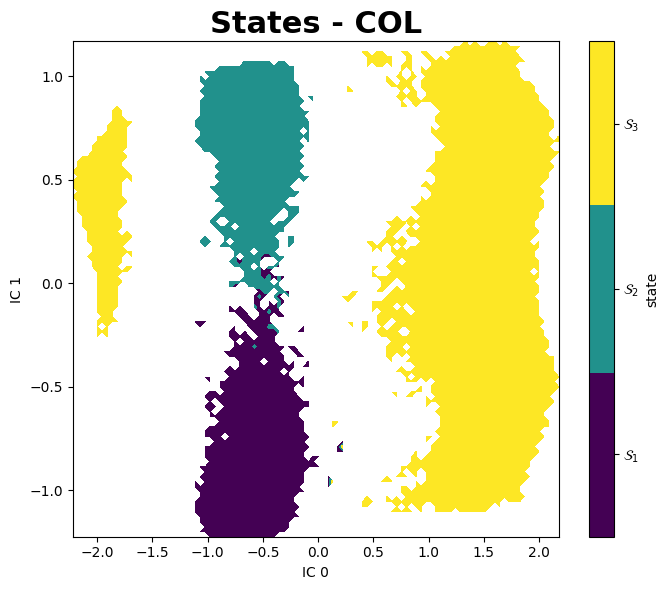
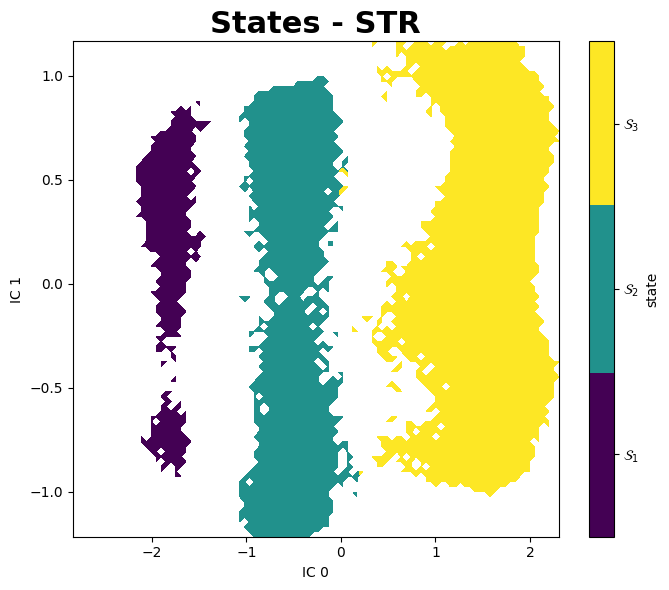
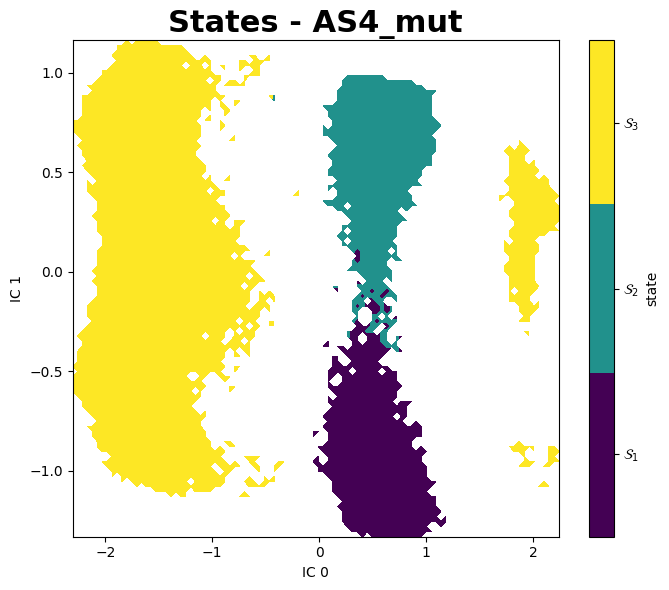
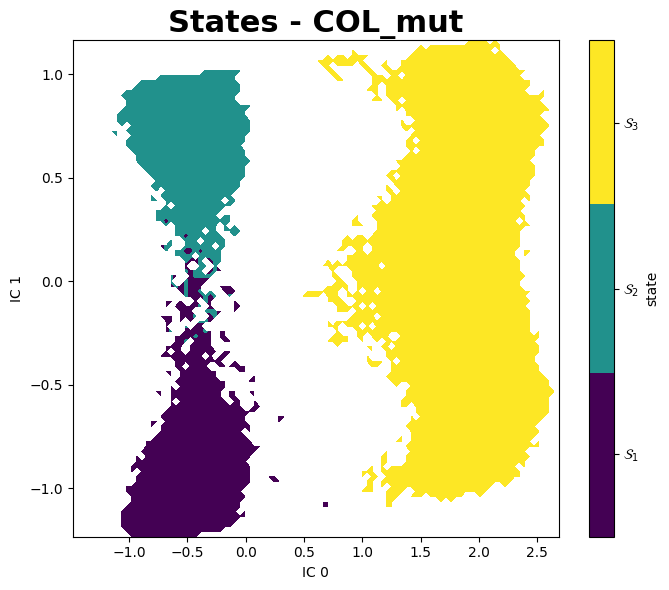
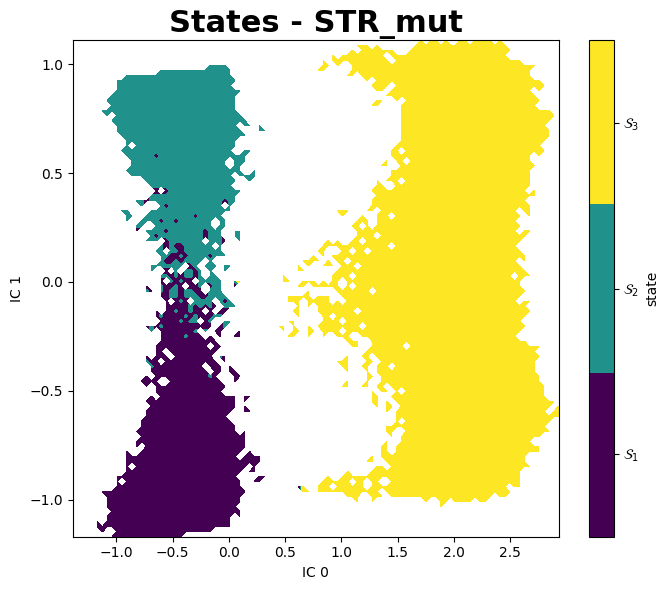
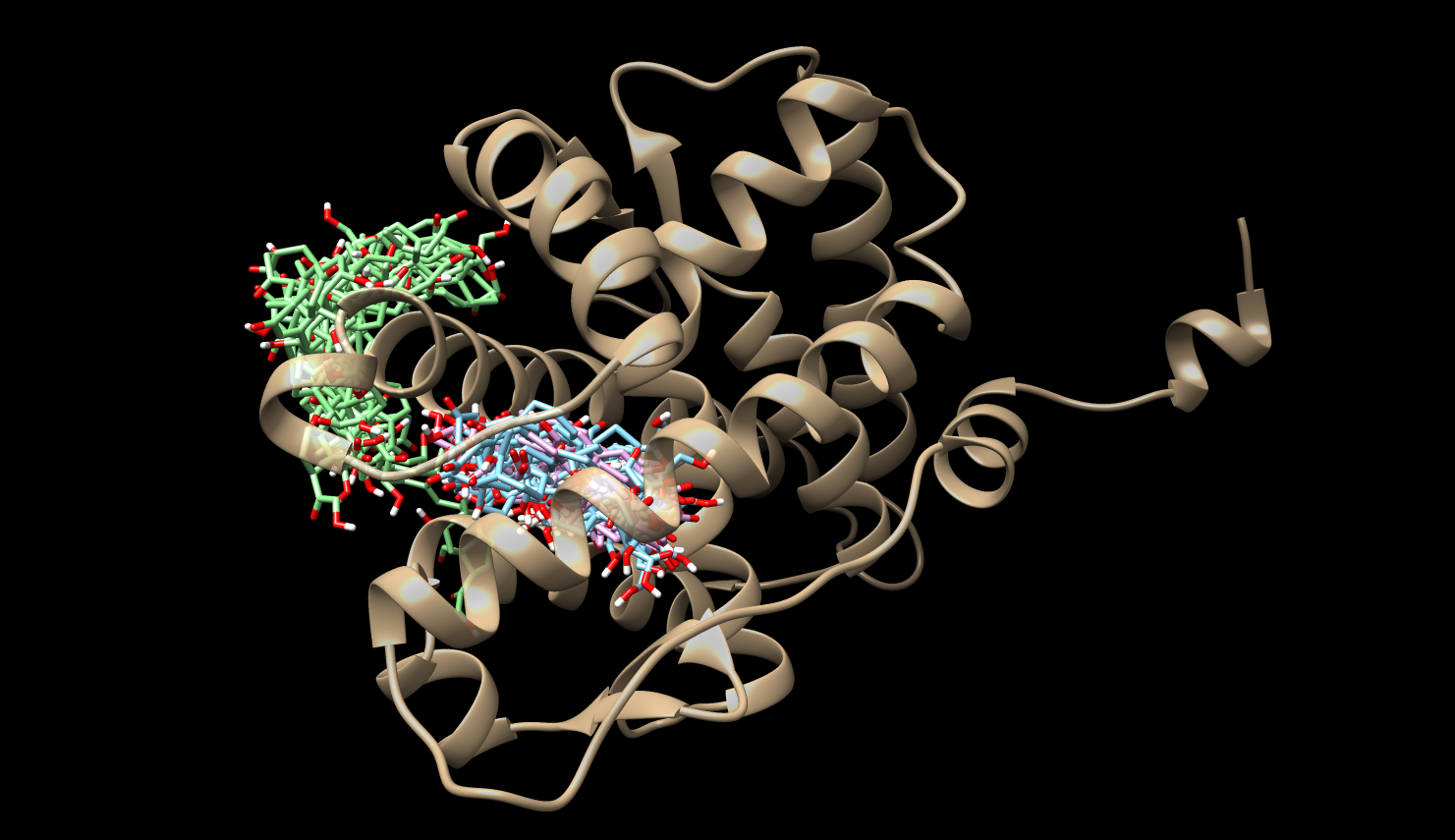
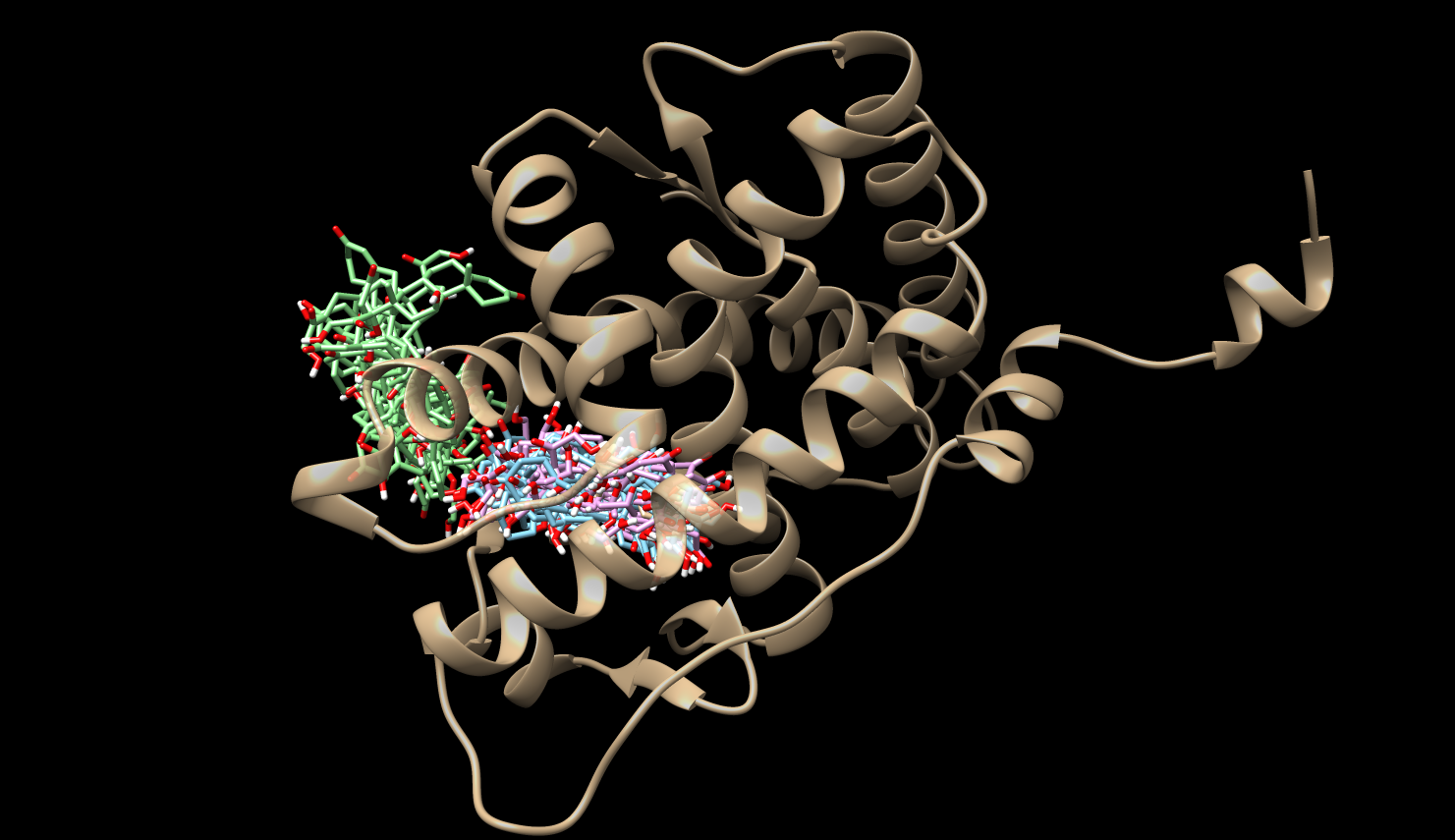
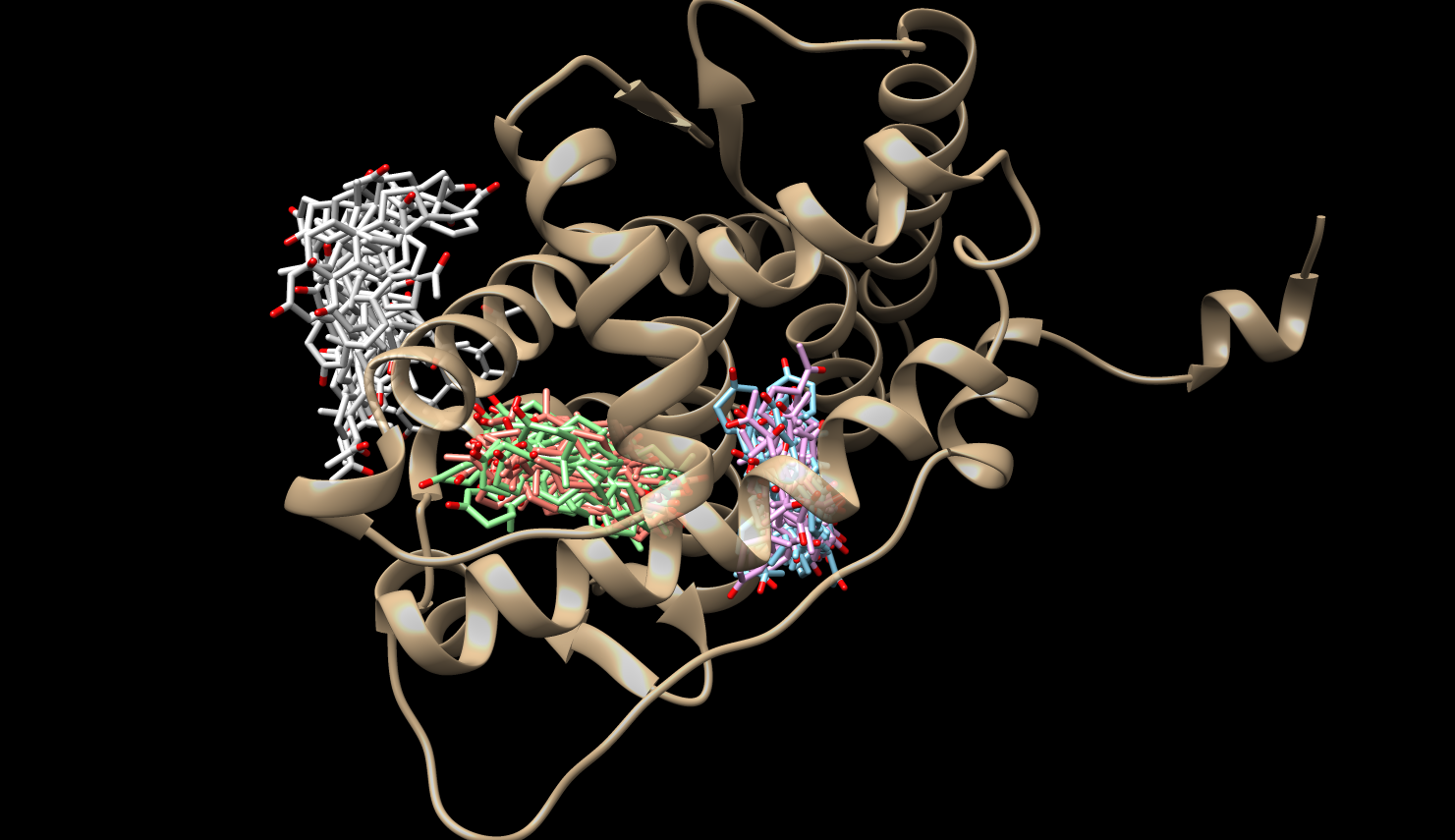
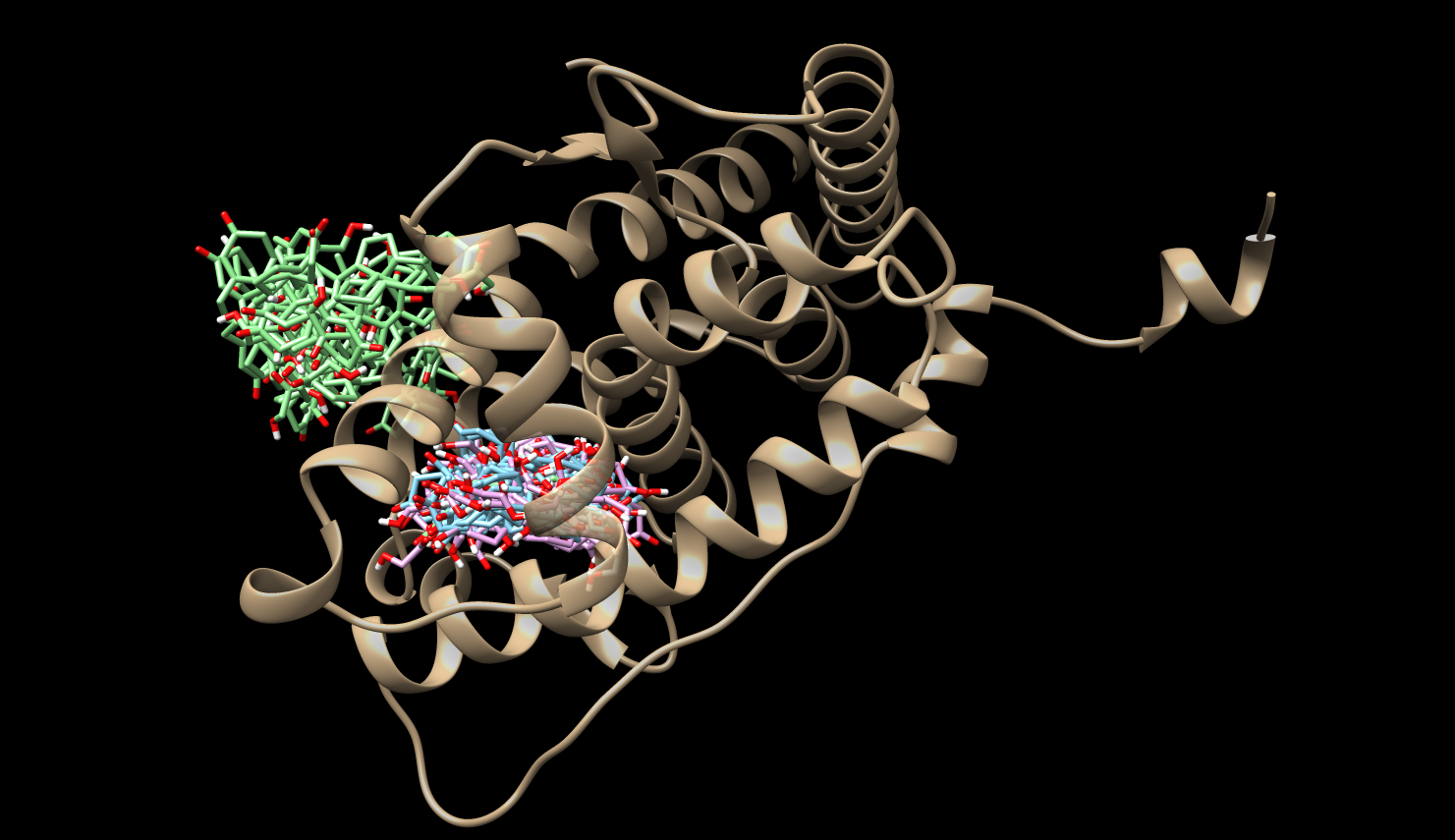
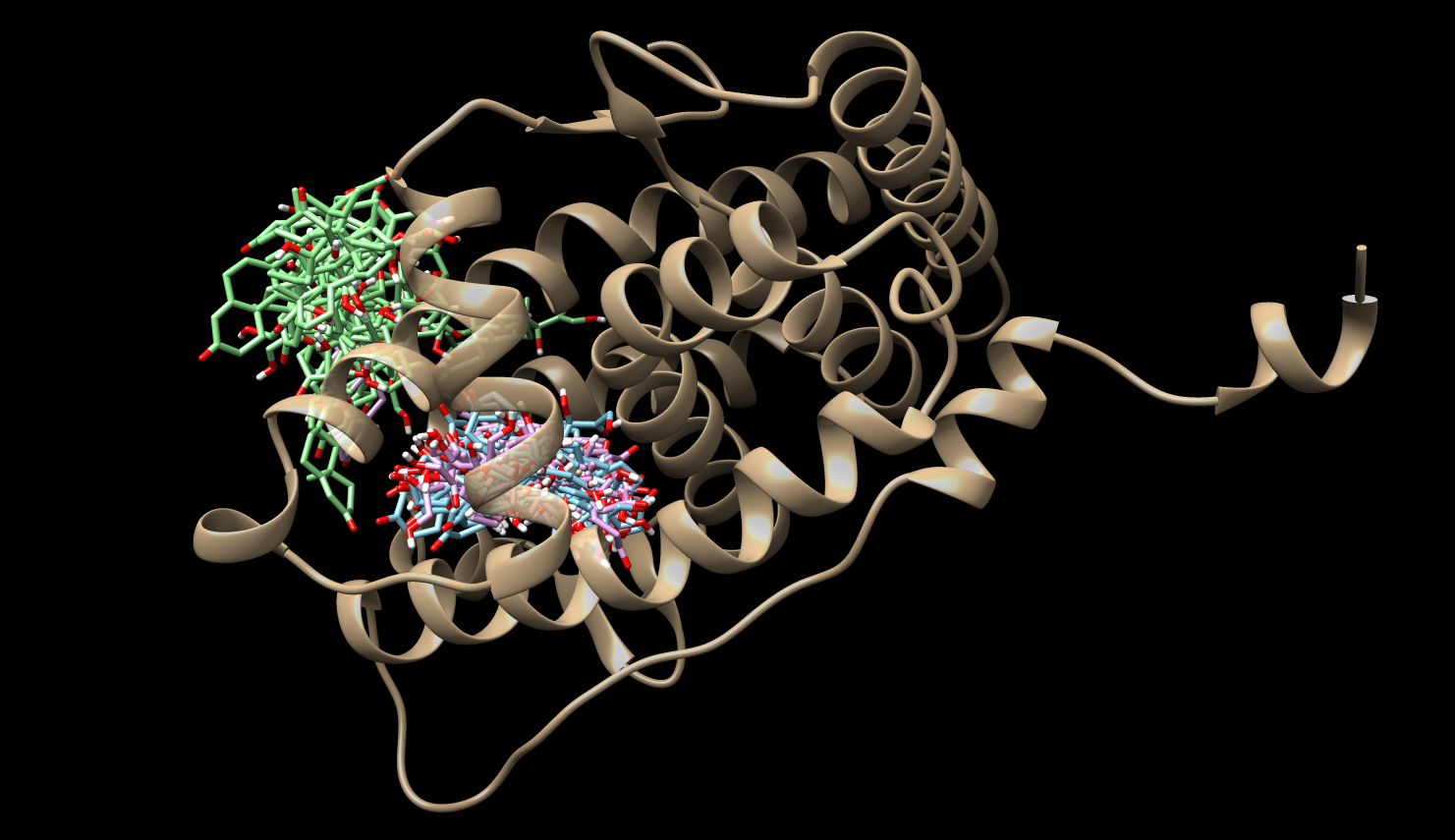
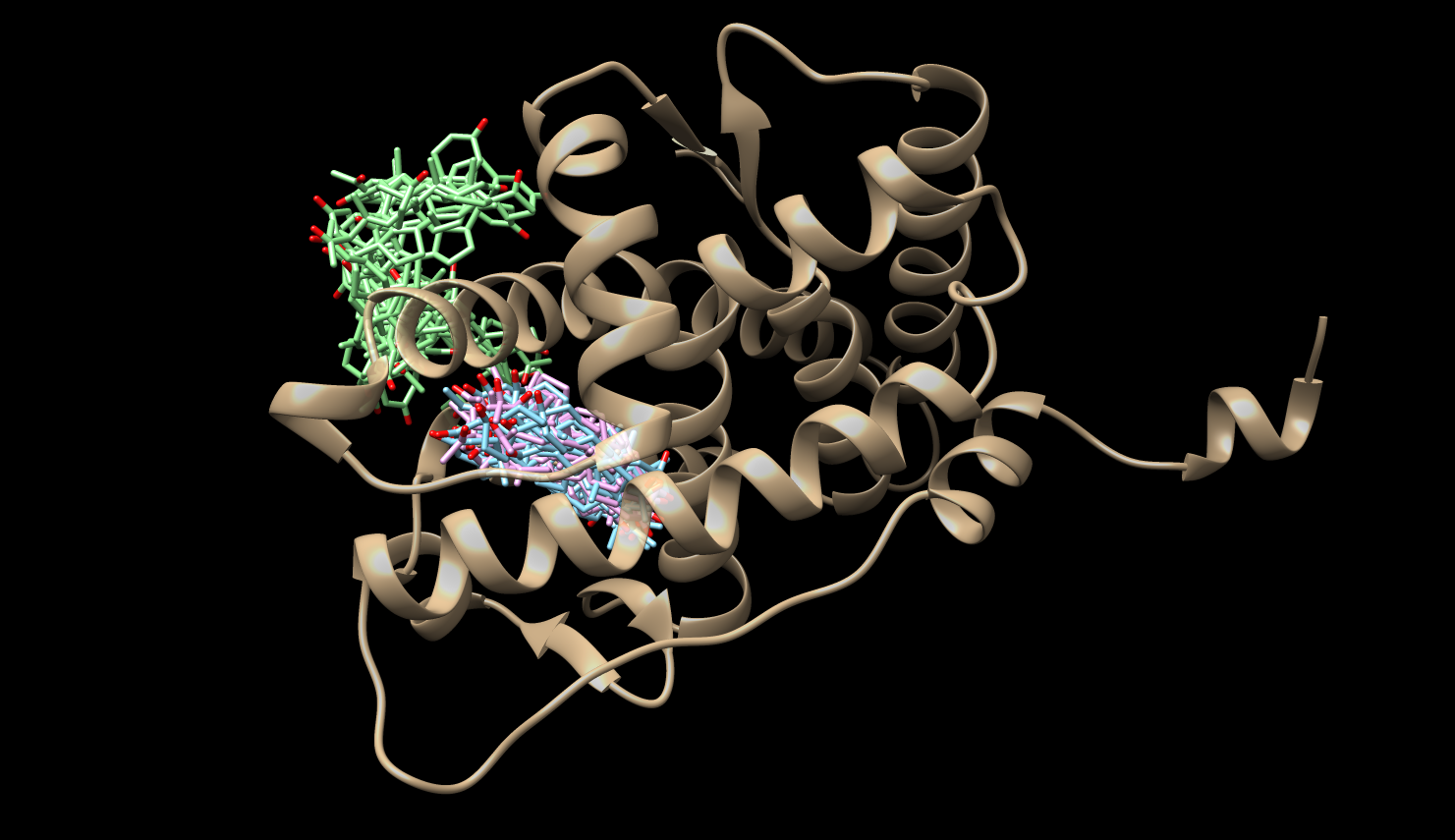
Mean First Passage Times (MFPTs)
All Sytems, All States
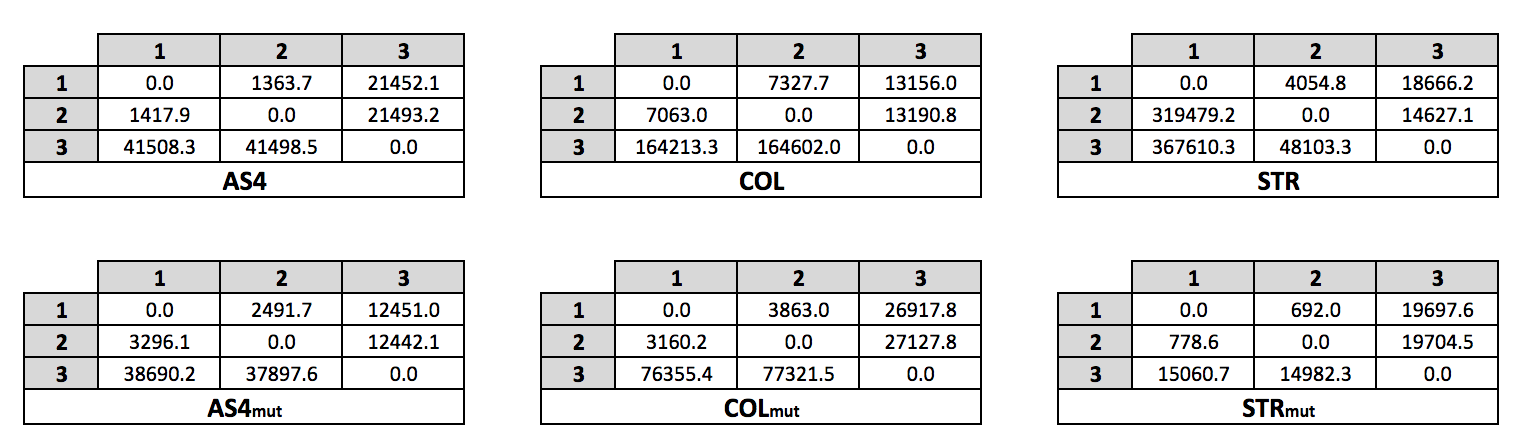
Bound State - Others
| Ligand | MFPT bound → other | MFPT other → bound |
|---|---|---|
| Aldo | 511.8 ± 8.3 | 34229.1 ± 5028.2 |
| Col | 512.9 ± 8.9 | 163482.9 ± 31149.2 |
| Str | 267.7 ± 3.3 | 57136.8 ± 8330.1 |
| Aldomut | 797.7 ± 16.4 | 38229.3 ± 5514.4 |
| Colmut | 968.2 ± 21.5 | 53617.9 ± 5090.8 |
| Strmut | 446.2 ± 5.5 | 9677.0 ± 1095.6 |